A US EPA grant will support testing of environmental monitoring protocols for measuring the outputs of AMR from wastewater treatment plants.
On 1 August 2024, the US Environmental Protection Agency (US EPA) awarded $2.37m to the Water Research Foundation, Virginia Tech, Arizona State University, West Virginia University, and the University of South Florida as part of a ~$3.6m consortium effort for a grant titled ‘Evaluation of Antimicrobial Resistance (AMR) in Wastewater and Sewage Sludge Treatment and Its Impact on the Environment’.
The research has four objectives:
Determine the ranges of mass loadings of antibiotic resistant bacteria (ARB) and resistance genes (ARGs) in wastewater treatment plant (WWTP) effluents and biosolids as a function of WWTP characteristics.
Estimate the degree to which WWTP effluent- and biosolid-borne ARB/ARGs attenuate or amplify in the environment and determine the controlling factors.
Evaluate evidence of WWTPs as a source of AMR infections in humans relative to other sources over a cross-section of US communities representative of diverse wastewater management scenarios.
Develop a human health risk assessment for WWTP effluent and biosolid sources of ARB/ARGs based on the knowledge gaps addressed through Objectives 1-3.
Engage and translate the research into actionable solutions for addressing AMR.
Wastewater treatment plants as a key barrier to the spread of deadly infections
Amy Pruden, University Distinguished Professor at Virginia Tech, and her collaborators have advanced foundational research over the last five years to develop the sampling and analysis methods and establish key collaborations with wastewater utilities, public health agencies, hospitals and health clinics, and medical researchers to set the stage for success with the US EPA grant that is now getting underway.
The research is scheduled to be completed in July 2027 and will provide quantitative insights into the contributions of wastewater to antibiotic-resistant pathogens and their ARGs to the water environment and ultimately into difficult-to-treat and deadly antibiotic-resistant infections in humans.
This will be achieved through a nationwide survey of surface water samples upstream and downstream from WWTP discharge points, along with culturing of Escherichia coli, Enterococcus spp. and Pseudomonas aeruginosa and the use of DNA sequencing technology to match environmental isolates and metagenome sequences to available clinical data.
Quantitative microbial risk assessment frameworks will be adapted to consider unique aspects of antibiotic resistance, including colonisation and delayed infection and the ability of pathogens to acquire ARGs from normal flora, in order to guide effective policy measures for stemming the spread of AMR.
This will include a selection of appropriate sewage sludge and wastewater treatment technologies that minimise the potential for the evolution and spread of AMR amongst microbial communities.
Another benefit of the effort will be the development and dissemination of culture, polymerase chain reaction (PCR), and DNA-sequencing-based protocols for monitoring AMR in environmental samples as a key connector across One Health sources and contributors to the spread of AMR in water.
Foundational to the success of the US EPA-supported effort will be recent research progress made by Pruden and her collaborators.
Towards standardised monitoring of AMR in water environments
Water is an essential thread that provides a pre-requisite for life: human, animal, and plant-like. Within a watershed, surface water bodies, including lakes, rivers, and streams, also receive point and non-point sources of pollution, including antimicrobials, antibiotics, faecal-indicator bacteria, pathogens, antibiotic-resistant bacteria, and ARGs.
Watersheds receive such pollutants from a variety of sources, including industrial discharge, WWTP effluents, and farm runoff. This makes surface water an ideal integrative monitoring point for One Health AMR monitoring.
However, sample collection and testing methods are needed to produce data that will be comparable across multiple water types (e.g., wastewater, river water) and also with other One Health-relevant matrices, such as clinical, food, livestock, and crop samples.
The Water Research Foundation initiated Project 5052 to meet this need and selected Professors Amy Pruden (Virginia Tech) and Valerie Harwood (University of South Florida) to lead up the research effort.
Through Project 5052, an overarching framework for monitoring AMR in water environments was developed, focusing on selecting the monitoring location and the method depending on the monitoring objective. They surveyed over 100 AMR experts across the One Health field and held a four-day expert workshop on developing this framework in May 2020.
This framework was published Open Access in the journal, Environmental Science & Technology1 and the overarching monitoring framework is shown in Fig. 1.
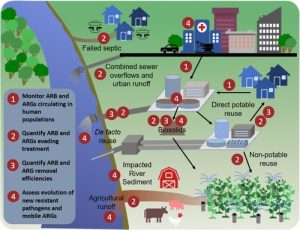
Standardising protocols for culture-based environmental AMR monitoring
Furthermore, as part of the Water Research Foundation Project 5052, they were able to carry out systematic literature reviews of candidate culture-based targets for AMR monitoring in water environments.
Culture-based targets present the advantage of verifying a viable target and also methods that can narrow in on specific bacteria of interest, such as faecal-indicator bacteria or pathogens.
Through a systematic literature review, they narrowed down targets and methods that are most promising for standardisation and for producing data that are comparable across general efforts to advance One Health monitoring of AMR in water, such as the World Health Organization TriCycle Protocol for ESBL E. coli.
A summary of promising methods for Enterococcus spp.2 is reported in Davis et al., and a summary of promising methods for Pseudomonas aeruginosa, Acinetobacter baumannii, and Aeromonas spp.3 is reported in Milligan et al.
Finally, they developed and validated a method for cefotaxime-resistant E. coli4 that adapts the US EPA Standard Method for E. coli with that of the TriCycle Protocol.
Standardising protocols for DNA-based environmental AMR monitoring
The advent of next-generation DNA sequencing technology circa 2010 has been a game-changer across numerous omics-related fields, including AMR.
DNA extracts of aquatic or other environmental samples can be directly extracted and subject to shot-gun sequencing in order to profile all of the thousands of ARGs representative of that sample, i.e., the “metagenome”. Pathogen gene markers and mobile genetic elements, such as plasmids, can also be annotated, providing important context about the ARGs.
For example, DNA sequencing data can be analysed to identify associations between ARGs and pathogens and to assess whether the ARG is on a mobile genetic element and, therefore, more likely to spread and be shared among different strains and species of bacteria. DNA sequencing information can also be uploaded to databases to support comparative analysis across One Health environments.
There is some hesitation in the adoption of metagenomics for broad-scale environmental monitoring; however, it is because of the cost involved, lack of standardised protocols, difficulty interpreting data, and high detection limits. To address these concerns, Pruden and her team conducted a systematic literature review to outline the steps towards standardisation of metagenomics for AMR monitoring5 and further validated an internal standard protocol to improve the quantitative capacity of metagenomics for this purpose, a method they dubbed as quantitative metagenomics.
Monitoring methods employing quantitative PCR droplet digital PCR can only target one or a few ARGs, mobile genetic elements, or pathogen markers at a time, but still retain value because of their relatively low detection limit and quantitative capacity. They conducted a systematic review of the application of qPCR for environmental AMR monitoring and extracted data from the literature to assess the range and variance of various types of ARGs found in various aquatic matrices6.
Bioinformatic tools to support DNA-based environmental AMR monitoring
Metagenomic sequencing data are rich with information, and multiple dimensions can be analysed to gain insight into AMR in the environment.
Of particular interest are the numbers and types of ARGs in a sample. These can then be further analysed to assess whether they encode resistance to antibiotics that are clinically important in human and/or veterinary medicine.
The ability to identify new ARG variants before they are known entities in public databases is also a concern. To this end, Pruden, Liqing Zhang (Virginia Tech) and their PhD student, Gustavo Arango Argoty, developed DeepARG7, which uses a deep learning approach to identify and classify ARGs according to their mechanisms and drug resistance class.
More recently, their post-doc Connor Brown led the development of a comprehensive, unified database of mobile genetic elements called mobileOG8, which can be used along with ARG databases to assess their mobility. mobileOG includes plasmids, transposons, integrons, and other mobile genetic elements and classifies them according to Hallmark genes or their orthologous groups, which avoids ambiguity in their annotation.
The MetaCompare pipeline packages deepARG and mobileOGdb together, applying an algorithm to compare metagenomes across environmental samples of interest in terms of their “resistome risk.” Resistome risk is defined as the degree to which ARGs in a metagenome are carried on the same strand of DNA as a mobile genetic element that is also annotated as being taxonomically related to known human pathogens.
The most recent MetaCompare pipeline9 differentiates Human Health Resistome Risk or the degree to which Rank I ARGs are mobile and carried within putative ESKAPEE pathogens, versus Ecological Resistome Risk, which is a broader algorithm that captures all ARGs and known human pathogens and assesses the general potential for AMR to spread to pathogens in a given environment.
Advancing wastewater-based surveillance policies to combat the spread of AMR
The COVID-19 pandemic elevated interest in wastewater-based surveillance of disease and rapidly expanded global infrastructure for this purpose.
At the same time, metagenomic sequencing was beginning to be applied to influent sewage to WWTPs, revealing striking patterns in ARGs and overall promise for wastewater-based surveillance of AMR. Amy Pruden, Peter Vikesland (Virginia Tech) and their team were funded by the US National Science Foundation through the Partnership for International Research and Education program from 2015-2023 for a project focused on using metagenomics to track ARGs through WWTPs at an international scale.

They noted striking patterns, with elevated relative abundances of ARGs in the sewage that we collected from two WWTPs in India, with the two WWTPS that were sampled in Sweden having the lowest relative abundances of total ARGs10.
Given that Sweden has the strictest regulations in place for preventing the spread of AMR across the One Health spectrum, the results suggest that metagenomic monitoring of this nature could provide a valuable approach to inform and assess policy and practice aimed at stemming the spread of AMR.
As part of this effort, Pruden and her collaborators subsequently launched a graduate student training program, also funded by the US National Science Foundation, focused on providing integrated student training in Data Science, Environmental Science and Engineering and Policy. The aim is to train a generation of future leaders capable of tackling complex challenges at Public Health, Environment, and Policy interfaces.
Her collaborators led a ‘Current Opinion in Microbiology’ article in 2021 laying out opportunities to harness wastewater-based surveillance of AMR to inform policy.
Additional works from this research group have further compared the metagenomes of human faeces versus sewage to gain insight into linkages between socioeconomics and AMR11.
Other studies from the team have made advances in estimating the extent to which a typical WWTP reduces ARGs12, the potential for antibiotics in the sewage13 to select for resistant bacteria during wastewater treatment14, and the extent to which advanced treatments for water reuse, such as advanced oxidation processes, can further reduce ARGs beyond conventional treatment15.
References
Liguori, K.; Keenum, I.M.; Davis, B.C.; Calarco, J.; Milligan, E.; Harwood, V.J.; and Pruden, A. (2022). Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control. Environmental Science and Technology. 56, 13, 9149–9160. https://doi.org/10.1021/acs.est.1c08918
Davis, B.C.; Keenum, I.M.; Calarco, J.; Liguori, K.; Milligan, E.; Pruden, A.; Harwood, V.J. (2022). Towards the standardization of Enterococcus culture methods for waterborne antibiotic resistance monitoring: A critical review of trends across studies. Water Research X. 17: 100161. https://doi.org/10.1016/j.wroa.2022.100161
Milligan, E.G.; Calarco, J.; Davis, B.C.; Keenum, I.M.; Liguori, K.; Pruden, A.; Harwood, V.J. (2023). A systematic review of culture-based methods for monitoring antibiotic resistant Acinetobacter, Aeromonas, and Pseudomonas as environmentally-relevant pathogens in wastewater and surface water. Current Environmental Health Reports. https://doi.org/10.1007/s40572-023-00393-9
Calarco, J.; Pruden, A.; Harwood, V.J. (2024). Comparison of Methods Proposed for Monitoring Cefotaxime-Resistant Escherichia coli in the Water Environment. Appl. Environ. Microbiol. Apr 4; 90(5): e02128-23. https://doi.org/10.1128/aem.02128-23
Davis, B.C.; Calarco, J.; Liguori, K.; Milligan, E.G.; Brown, C.L.; Gupta, S.; Harwood, V.J.; Pruden, A.; Keenum, I.M. (2023). Recommendations for the use of Metagenomics for Routine Monitoring of Antibiotic Resistance in Wastewater and Impacted Aquatic Environments. Current Opinion in Environmental Science and Technology https://doi.org/10.1080/10643389.2023.2181620
Keenum, I.; Liguori, K.; Calarco, J.; Davis, B.C.; Milligan, E.; Harwood, V.J.; and Pruden, A. (2022). A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater. Critical Reviews in Environmental Science and Technology, 25 pages. https://doi:10.1080/10643389.2021.2024739
Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.J.; Zhang, L. (2018). DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 6(1):23 https://doi.org/10.1186/s40168-018-0401-z
Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.M.; Vikesland, P.J.; Pruden, A.P.; Zhang, L. (2022) mobileOG-db: a manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. https://doi.org/10.1128/aem.00991-22
Afrin Rumi, M.A., Oh, M., Davis, B.C., Juvekar, A., Brown, C.L., Vikesland, P.J., Pruden, A., Zhang, L. (2024). MetaCompare 2.0: Differential ranking of ecological and human health resistome risks. FEMS Microbiol Ecol Volume 100, Issue 12, December 2024, fiae155, https://doi.org/10.1093/femsec/fiae155
Prieto Riquelme, M.V.; Garner, E.D.; Gupta, S.; Metch J.; Zhu, N.; Blair, M.F.; Arango-Argoty, G.; Maile-Moskowitz, A.; Li, A-D.; Flach, C-F.; Aga, D.S.; Nambi, I.M.; Larsson, D.G.J.; Bürgmann, H.; Zhang, T.; Pruden, A.; and Vikesland, P.J. (2022). Demonstrating a Comprehensive Wastewater-Based Surveillance Approach That Differentiates Globally Sourced Resistomes. Environ. Sci. Technol. 56, 21, 14982–14993 https://doi.org/10.1021/acs.est.1c08673
Gupta, S., Wu, X., Pruden, A., Zhang, L., Vikesland, P.J. (2024). Global scale exploration of human faecal and sewage resistomes as a function of socio-economic status. Nature Water 2, 975–987 (2024). https://doi.org/10.1038/s44221-024-00310-w
Garner, E.; Maile-Moskowitz, A.M.M.; Angeles, L.F.; Flach, C-F; Aga, D.S.; Nambi, I.; Larsson, D.G.J.; Bürgmann, H.; Zhang, T.; Vikesland, P.J.; and Pruden, A. (2024). Metagenomic Profiling of Internationally Sourced Sewage Influents and Effluents Yields Insight into Selecting Targets for Antibiotic Resistance Monitoring. Environmental Science & Technology. 58, 37, 16547-16559 https://doi.org/10.1021/acs.est.4c03726
Brown, C.L., Maile-Moskowitz, A., Lopatkin, A.J., Xia, K., Logan, L., Davis, B.C., Zhang, L., Vikesland, P.J., Pruden, A. (2024). Selection and horizontal gene transfer underlie microdiversity-level heterogeneity in resistance gene fate during wastewater treatment. Nature Communications. 15, 5412 https://doi.org/10.1038/s41467-024-49742-8
Keenum, I.; Calarco, J.; Majeed, H.; Hager-Soto, E.E.; Bott, C.; Garner, E.D.; Harwood, V.J.; Pruden, A. (2024). To what extent do water reuse treatments reduce antibiotic resistance indicators? A comparison of two full-scale systems. Water Research. 254: https://doi.org/10.1016/j.watres.2024.121425
Source link

