The dealii-X project aims to create highly accurate digital twins of human organs using advanced computational models, enhancing insights into biomechanics and improving medical understanding and treatment of diseases.
The project “dealii-X: an Exascale Framework for Digital Twins of the Human Body” is one of the EuroHPC Centres of Excellence, aiming to develop a scalable, high-performance computational platform to create accurate digital twins of human organs. The project has a strong mathematical component and builds on the deal.II library,¹ a toolbox for enabling the rapid development of finite element models for the numerical approximation of the solution of partial differential equations. These models have traditionally been used to describe a wide variety of engineering problems, be it how heat spreads through a device, how a bridge bends under load, or how sound moves through the air, all of which are solved by representing the underlying laws of physics on a computer. The key idea is to break a complicated shape into many small, simple pieces (elements), approximate the solution within each piece, and then stitch those local approximations together. Even though the solution approximation on each element is made up by linear relationships with a few unknown coefficients, the orchestration through an ensemble of a large number of elements makes approximations very accurate. This approach turns hard continuous equations into large but manageable systems of linear algebra. The dealii-X project tries to carry over this success to biomechanics, more precisely, to creating accurate models for organs in the human body – lungs, heart and cardiovascular system, brain, liver, and cell mobility processes.
While computational models of various kinds are not new to biomedicine, most of them were still limited in their capabilities, typically aiming at simplified settings with limited quality.
The steady increase in computing capabilities and progress in mathematical and mechanical modelling has opened the door to perform ground-up models that closely mimic the processes in the organs that every one of us has in their body. These digital twins provide new insights into medicine and potential treatment of diseases mankind suffers from today, with the first attempts going into the understanding of the main drivers in organs.
Mathematically, the models are challenging to set up: one difficulty is that the configurations are different for every person and even undergo continuous remodelling throughout a human’s lifetime, necessitating careful collection of data to actually create the geometries to be modelled, and yet leaving many aspects uncertain. On top of that, to accurately model the organs, a wide range of scales needs to be considered, starting from the formation of cells to whole tissue layers, where different mechanical processes like elastic deformation of strongly anisotropic hyperelastic materials are interacting with the flow of blood, a mixture of plasma and blood cells, or the flow of air. The dealii-X project leverages exascale computing capabilities to drive models of these complex biological processes at an unprecedented level of detail. The project formally started in October 2024, and the first results started to line up at this point. This article provides insight into four selected models developed in the dealii-X consortium, where researchers located in Germany, Italy, France, and Belgium strive to deliver application excellence with new models.
Respiratory mechanics
Mechanical ventilation is crucial for patients with impaired pulmonary function. Clinicians must balance effective respiration with strategies that protect lung tissue, because mechanical ventilation can cause ventilator-induced lung injury. Unfortunately, the local effects of mechanical ventilation are challenging to measure or observe by medical imaging. Computational models promise physics-based forecasts on the impact of different ventilation maneuvers on patient-specific geometries.
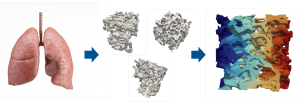
At present, most models fail to incorporate many fundamental effects in respiratory mechanics; surfactant being among the most notable. Surfactant molecules in the fluid lining of the alveoli reduce surface tension, thereby increasing lung compliance. To understand surfactant effects and integrate them into patient-specific reduced-order or homogenised models, we require numerous highly resolved simulations of delicate and complex alveolar structures.² A team at Technical University of Munich, Germany, centered around Professor Wolfgang A. Wall and Buğrahan Temür, performs these simulations using the open-source ExaDG software project. ExaDG is based on the deal.II library and provides highly efficient solvers for problems in fluid mechanics, solid mechanics, and fluid-structure-interaction. The extensible software architecture enables the team to implement surfactant dynamics³ in the matrix-free hyperelasticity solver.⁴ The exceptional performance allows the researchers to handle the high resolution and the large number of different setups required to extract information from the fine-scale alveolar structure simulations.
Matrix-free cardiovascular simulations
Cardiovascular diseases are the leading cause of death worldwide, making it essential to understand cardiac function and blood circulation. Cardiovascular modelling provides a powerful framework to study blood flow, heart mechanics, and tissue perfusion, supporting improved diagnosis and treatment.
High-fidelity 3D cardiac models⁵ require substantial computational resources, as they involve complex interactions between blood flow, tissue mechanics, and biochemistry. These resources are usually found in high-performance parallel computing environments.⁶ Therefore, effective cardiac simulations require not only physically accurate mathematical models, but also efficient and scalable algorithms and software that can leverage the capabilities of modern computing hardware.

Within dealii-X, Professor Luca Dede’ with the researchers Michele Bucelli, Andrea Tonini, and Junxiang Wang at Politecnico at Politecnico di Milano, Italy, introduce a matrix-free computational framework for efficient large-scale cardiovascular simulations. An example simulation is shown in Fig. 2. Unlike conventional matrix-based approaches, which precompute and store large system matrices and often under-utilise computing hardware, the matrix-free approach computes operations on the fly, significantly reducing memory usage and communication overhead. The improvements the team gets in run-time are the result of a thorough process of various partners in dealii-X in aligning algorithms with the capabilities of modern high-performance computing systems. The team investigates the impact of matrix-free algorithms on cardiocirculatory simulations,⁷ focusing on their effects on computational efficiency and resource usage. These algorithms have the potential to enable high-resolution, patient-specific digital heart models that may help researchers and clinicians better understand cardiovascular diseases, explore treatment options, and support progress toward more personalised and data-driven cardiovascular care.
Gaining new insights into the human brain by inverse modelling of tissue mechanics
Understanding and predicting the biomechanical properties of human brain tissue can improve the diagnosis and treatment of pathological conditions. Thereby, predictions based on finite element models can assist in preoperative planning and facilitate surgical procedures, causing minimal damage. Our nonlinear poro-viscoelastic model resembles the biphasic nature of brain tissue and captures its characteristic mechanical behavior. The model improves our understanding of the interactions between the solid tissue matrix and free-flowing interstitial fluid. Once the brain region-specific material model parameters are reliably calibrated through inverse parameter identification schemes from experimental data, they are applied to patient-specific full brain models generated from MRI data. The deal.II-based open-source project ExaBrain, developed by Professor Silvia Budday and Alexander Greiner at Friedrich-Alexander-University of Erlangen-Nuremberg in Germany, aims to enable both, parameter identification (many small-scale simulations) and full brain applications (few large-scale simulations), exploiting state of the art high performance computing methods. This includes specialised direct and iterative solvers and potentially matrix-free methods to increase accuracy using higher-order finite elements.
At the foundation of human cells
Single and collective cell motility, the fundamental mechanisms behind tumour metastasis and embryogenesis, arise from a complex and uninterrupted sequence of polymerisation and depolymerisation of actin. The interactions between receptors on the cell lipid membrane and ligands in the extracellular matrix are essential to this end, as well as to several other physiological and pathological events. Fig. 4 below illustrates the numerical and experimental evolution in time of the relocation of Vascular Endothelial Growth Factor Receptors on the advecting membrane. Those interactions ultimately yield the reorganisation of the cytoskeleton, i.e., the creation of a new protrusive network at the leading edge of cells and the contraction of its rear through the myosin action in the stress fibers. The team by Professor Alberto Salvadori at the Università degli Studi di Brescia, Italy, develops and implements novel continuum multiphysics partial differential equations that govern cellular motility:⁸ simulations in Fig. 4a were not provided with these tools, and protrusion was imposed via external forces. The team is working on a new software to complement and advance the pre-exascale deal.II suite currently available at The Mechanobiology Research Center, UNIBS, to enable cellular investigations that are interconnected in terms of protein relocation and fluid-cell interactions to the organ modelisation codes developed in the framework of the research. Ultimately, the goal is to facilitate mechanobiological research and identify therapies that will limit the spread of harmful cells while increasing the motility of beneficial ones. Codes will be verified, and models will be validated against in vitro experimental campaigns.
Summary
These examples illustrate the wide variety of models and development within dealii-X. While the processes in a heart, a lung, or the brain are very different, the mathematical picture shows many similarities and allows researchers in the project consortium to enhance the models as a collective. Using large-scale computers pushes the boundary of what is possible and builds the foundation for exciting discoveries. Not the least, many of the consortium members are also passionate about what drives the large-scale parallel computers of the current and future generation, and to find ways to make the best use of the potential in these architectures. The solutions will immediately help the biomedical applications, but many of them are generically applicable and will be integrated into the base mathematical library deal.II, which then also improves applications in engineering, earth sciences, and many of the fields beyond the activities in dealii-X, just as dealii-X can learn from related disciplines in a true multi-disciplinary environment.
References
https://dealii.org
S. M. K. Rausch, D. Haberthür, M. Stampanoni, J. C. Schittny, and W. A. Wall. “Local Strain Distribution in Real Three-Dimensional Alveolar Geometries”. In: Annals of Biomedical Engineering 39.11 (2011), pp. 2835–2843.
L. Wiechert, R. Metzke, and W. A. Wall. “Modeling the Mechanical Behavior of Lung Tissue at the Microlevel”. In: Journal of Engineering Mechanics 135.5 (2009), pp. 434–438.
R. Schussnig, N. Fehn, P. Munch and M. Kronbichler. “Matrix-Free Higher-Order Finite Element Methods for Hyperelasticity”. In: Computer Methods in Applied Mechanics and Engineering 435 (2025), p. 117600.
M. Fedele, R. Piersanti, F. Regazzoni, M. Salvador, P. C. Africa, M. Bucelli, et al. and A. Quarteroni. “A comprehensive and biophysically detailed computational model of the whole human heart electromechanics”. In: Computer Methods in Applied Mechanics and Engineering 410 (2023), p. 115983.
M. Bucelli. “The lifex library version 2.0”. In: ACM Transactions on Mathematical Software (2024).
P. C. Africa, M. Salvador, P. Gervasio, L. Dede, and A. Quarteroni. “A matrix–free high–order solver for the numerical solution of cardiac electrophysiology”. In: Journal of Computational Physics 478 (2023), p. 111984.
M. Serpelloni, M. Arricca, C. Ravelli, E. Grillo, S. Mitola, A. Salvadori (2023), Mechanobiology of the relocation of proteins in advecting cells: modeling, experiments, and simulations, Biomechanics and Modeling in Mechanobiology, 22:1267–1287
A. Salvadori, C. Bonanno, M. Serpelloni, R.M. McMeeking, (2024), On the generation of force required for actin-based motility., Scientific Reports, 14:18384
Authors
Martin Kronbichler (coordinator)
Silvia Budday
Luca Dede’
Alberto Salvadori
Wolfgang A. Wall
Please Note: This is a Commercial Profile
Please note, this article will also appear in the 25th edition of our quarterly publication.
Source link

